The aim of this tutorial is to briefly present an example of an OPES_METAD simulation, together with some useful post-processing tools for obtaining an estimate of the free energy surface. This tutorial does not contain theory explanations. The OPES method is presented in detail in Ref. [68] and in its supporting information.
The TARBALL for this project contains the following:
FES_from_Reweighting.py: script for estimating free energy surface via reweightingFES_from_State.py: analogous to sum_hills, but for OPES simulationsState_from_Kernels.py: script for obtaining an OPES state file from a kernels fileWe will perform Langevin dynamics on the following two-dimensional model potential:
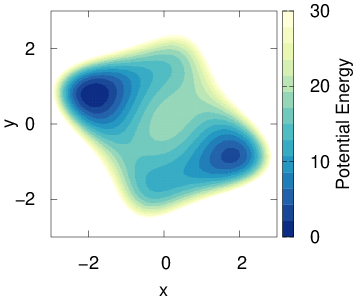
To run such simulations, we use the pesmd tool and define the potential directly inside the plumed input file.
We choose as collective variable (CV) the \(x\) coordinate only, in order to mimic a realistic situation in which some of the slow modes of the system are unknown and not accelerated by the biasing. The following is a typical plumed input for performing an OPES_METAD simulation:

UNITSNATURALp: DISTANCE( default=off ) use natural unitsATOMS=1,2the pair of atom that we are calculating the distance between.COMPONENTSopes: OPES_METAD( default=off ) calculate the x, y and z components of the distance separately and store them as label.x,ARG=p.xthe input for this action is the scalar output from one or more other actions.TEMP=1compulsory keyword ( default=-1 ) temperature.PACE=500compulsory keyword the frequency for kernel depositionBARRIER=10 PRINTcompulsory keyword the free energy barrier to be overcome.FILE=COLVARthe name of the file on which to output these quantitiesSTRIDE=500compulsory keyword ( default=1 ) the frequency with which the quantities of interest should be outputARG=p.x,p.y,opes.biasthe input for this action is the scalar output from one or more other actions.
By default an OPES_METAD simulation generates a KERNELS file that, similarly to the HILLS file from METAD, contains all the Gaussians deposited during the run, and can be used for restarting a simulation. We also print a COLVAR file containing the system coordinates and the instantaneous value of the bias. Here is the time evolution of the \(x\) coordinate.
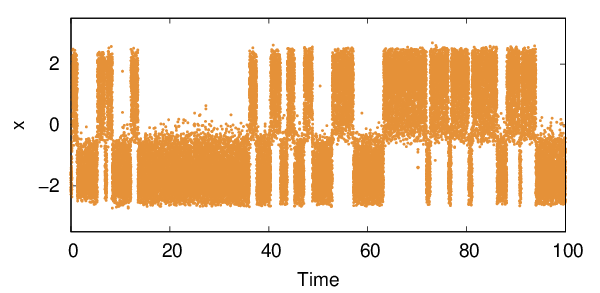
From this simulation we can estimate via reweighting the free energy surface (FES) along \(x\). We use a weighted kernel density estimation with bandwidth sigma, and use block average for uncertainty estimation. You should run the following script from the same folder where the COLVAR file is stored.
../FES_from_Reweighting.py --kt 1 --sigma 0.03 --blocks 5
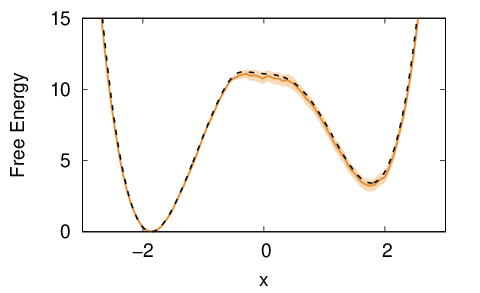
As value for sigma we used the one from the last line of the KERNELS file. Notice that a similar FES estimate could have been obtained using REWEIGHT_BIAS instead of the python script. For this simple system we can compare the result with the true FES (black dashed line):
logweight column as biasWe now run a well-tempered metadynamics simulation on the same system, to get an idea of the differences between the two methods. This is the plumed input used:

UNITSNATURALp: DISTANCE( default=off ) use natural unitsATOMS=1,2the pair of atom that we are calculating the distance between.COMPONENTSmetad: METAD ...( default=off ) calculate the x, y and z components of the distance separately and store them as label.x,ARG=p.xthe input for this action is the scalar output from one or more other actions.TEMP=1the system temperature - this is only needed if you are doing well-tempered metadynamicsPACE=500compulsory keyword the frequency for hill additionHEIGHT=1the heights of the Gaussian hills.SIGMA=0.185815compulsory keyword the widths of the Gaussian hillsBIASFACTOR=10use well tempered metadynamics and use this bias factor.GRID_MIN=-3.5the lower bounds for the gridGRID_MAX=3.5the upper bounds for the gridGRID_BIN=200the number of bins for the gridCALC_RCT... PRINT( default=off ) calculate the \f$c(t)\f$ reweighting factor and use that to obtain the normalized bias [rbias=bias-rct].ThisFILE=COLVARthe name of the file on which to output these quantitiesSTRIDE=500compulsory keyword ( default=1 ) the frequency with which the quantities of interest should be outputARG=p.x,p.y,metad.rbias,metad.rctthe input for this action is the scalar output from one or more other actions.
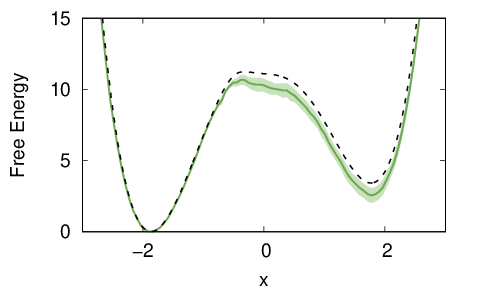
And here is the resulting \(x\) trajectory.

One can see that with metadynamics more transitions are present, especially in the initial part of the simulation. This is because the bias is changing quickly and can efficiently push the system through high free-energy pathways. Unfortunately, these out-of-equilibrium transitions cannot be efficiently reweighted and might lead to a systematic error.
To avoid this systematic error, one must remove the part of the metadynamics simulation where the bias is not quasi-static. You can do so using the option --skiprows in the FES_from_Reweighting.py script. Removing an initial transient might be useful also for OPES simulations, in particular when the chosen BARRIER is much higher than the real one.
A quick way to get a FES estimate along the biased CVs in OPES, is to directly look at the instantaneous bias \(V_t(x)\), since at convergence \(F(x)=-(1-1/\gamma)^{-1}V(x)\). For metadynamics simulations one should use the rbias instead, which is the bias shifted by the \(c(t)\) factor. In Fig. 6 we plot the COLVAR file using on the x-axis the CV \(x\) and on the y-axis this instantaneous FES estimate, \(F_t(x)\). Points are colored according to the simulation time.
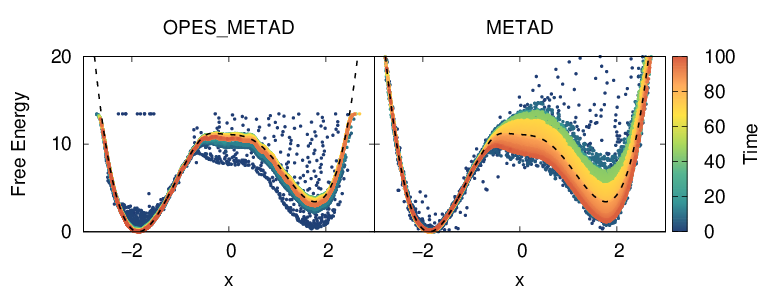
We can see that the running estimate from metadynamics has much broader oscillations. This can be seen also in Fig. 3 of Ref. [68], where only the free energy difference between the basins is plotted. It is also clearly visible that the OPES simulation does not explore free energy regions much higher than the given BARRIER parameter.
Another more common way of obtaining this direct FES estimate from the metadynamics bias is via the sum_hills tool, e.g. with the following command:
plumed sum_hills --hills HILLS --mintozero --stride 1000
An analogous procedure can be done also with OPES, starting from the STATE file printed during the simulation. This file contains the compressed kernels that are used internally to define the bias, together with all the information needed for a smooth restart. The syntax to print it is similar to the one used to print the bias grid in METAD :

UNITSNATURALp: DISTANCE( default=off ) use natural unitsATOMS=1,2the pair of atom that we are calculating the distance between.COMPONENTSopes: OPES_METAD ...( default=off ) calculate the x, y and z components of the distance separately and store them as label.x,ARG=p.xthe input for this action is the scalar output from one or more other actions.TEMP=1compulsory keyword ( default=-1 ) temperature.PACE=500compulsory keyword the frequency for kernel depositionBARRIER=10compulsory keyword the free energy barrier to be overcome.STATE_WFILE=STATEwrite to this file the compressed kernels and all the info needed to RESTART the simulationSTATE_WSTRIDE=500*1000number of MD steps between writing the STATE_WFILE.STORE_STATES...( default=off ) append to STATE_WFILE instead of ovewriting it each time
If you did not dump the STATE file while running your simulation, you can recover it with the driver (see READ). The script State_from_Kernels.py contains the minimal input to do so, and only requires the KERNELS file.
If the STATE file is available, one can run the provided script with the following command:
../FES_from_State.py --kt 1 --all_stored
For OPES_METAD the resulting FES estimate will be very similar to the one obtained from reweighting.
In this tutorial a single simulation is presented for OPES and metadynamics, but you can check Fig. S2 and S3 in the supporting information of Ref. [68] to see the results obtained by averaging 100 independent realizations.
The OPES_METAD has a few quantities one can print to monitor the simulation:
rct, contrary to the one of METAD, converges to a constant and can be useful to quickly check the status of the simulation, especially when running with many CVs. It should not be used for reweighting.zed should change only when a new region of CV-space is sampled.neff is an estimate of the effective sample size and should always grow.nker is the number of compressed kernels. If it is equal to the number of deposited kernels, it means that the simulation never comes back to where it has already been (too many CVs? too small SIGMA? too small COMPRESSION_THRESHOLD?).Finally, here are some optional keywords that might be useful:
NLIST activates a neighbor list over the kernels. It can considerably speed up the simulation, especially when using two or more CVs.SIGMA_MIN can be useful to reduce the total number of compressed kernels (nker) and thus speed up the simulation. It can help especially with long simulations, and can also be added after a restart (see also FIXED_SIGMA).